Research Interests
Design and Synthesis of Molecules with Biological Significance
Research interests in our laboratory are at the interface of chemistry and biology. The focus is on structure‐based design and synthesis of small molecules that can modulate essential pathways of infectious disease organisms with an emphasis on small molecule–protein interactions and target specificity. Our laboratory utilizes fragment‐based screening, de novo design, the design and synthesis of versatile fragment building blocks from natural products and construction of fragment libraries to explore enzyme selectivity.
Specific areas of interest include:
- Fragment‐based design of novel small‐molecules to selectively disrupt key enzymatic interactions in essential enzymatic pathways;
- Design and synthesis of novel small‐molecules to selectively inhibit the phosphodiesterase type 4 (PDE4) enzymes;
- Natural product isolation and characterization. Our group uses a multidisciplinary approach toward achieving these research goals including synthetic organic chemistry, computer modeling, protein crystallography, molecular and cell biology.
Fragment‐based Design of Novel Small‐molecules to Selectively Disrupt Key Enzymatic Interactions in Essential Enzymatic Pathways
Fragment based drug discovery is a rapidly growing technique in medicinal chemistry, that utilizes protein crystallography to discover unique fragments that bind to protein targets of biological interest. These low molecular weight fragments can then be optimized using medicinal chemistry techniques to obtain new compounds with low nM potencies. This can be achieved with a limited number of compounds, especially if good structural data is present. These techniques are being applied to enzymes in the non‐mevalonate isoprenoid biosynthetic pathway. We have established collaborations with research groups that have excellent structural capabilities and experience.
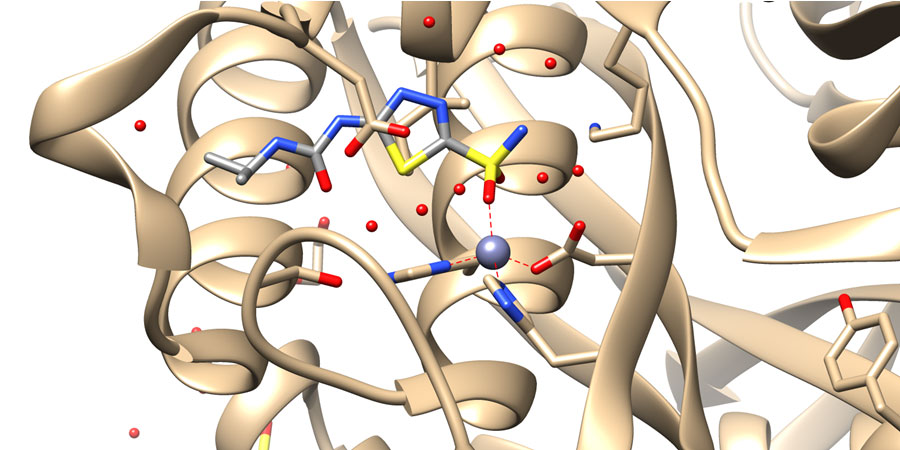
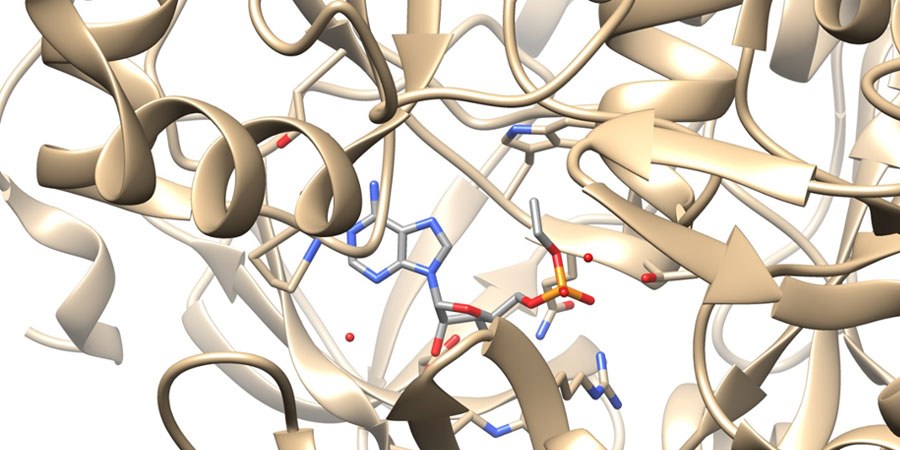
Enzymes such as Acetyl-coenzyme A synthetase are essential to microorganisms such as the pathogenic fungi Cryptococcus neoformans, Coccidioides immitis. Inhibitors of this enzyme have potential for a new class of anti-fungal drugs. A fragment based approach to finding inhibitors of this enzyme is being pursued. We have collaborations with Dr. Damian Krysan from the University of Iowa and the Seattle Structural Genomics Center for Infectious Disease (SSGCID) which has excellent structural capabilities and experience.
Contact Us
Faraday Hall 350Northern Illinois University
DeKalb, IL 60115
Phone: 815-753-1463
Fax: 815-753-2902
Email: thagen@niu.edu
Mailing Address
Timothy J. Hagen, Ph.D.Department of Chemistry and Biochemistry
Northern Illinois University
1425 W. Lincoln Hwy.
DeKalb, IL 60115‐2828
We frequently use Fragment-Based Drug Discovery (FBDD) for lead finding and lead optimization in ongoing collaborative projects with the Centers for Infectious Disease (CID), Dr. Damian Krysan University of Iowa and James Horn at NIU. These collaborators have provided purified enzymes for protein ligand studies. We have utilized Saturation Transfer Difference (STD) NMR techniques to determine epitope mapping for ligands binding to proteins and NMR to determine dissociation constants (KD) values. These NMR methods are very valuable techniques used to study protein-ligand interactions, binding affinity, and lead optimization in medicinal chemistry. These NMR experiments provide an excellent training opportunity for upper-level undergraduate students and graduate students. Initial training in these techniques begins with epitope mapping and binding affinity determination for tryptophan analogs to Human Serum Albumin (HSA), then students advance to other proteins and ligands. These techniques are not only is by medicinal comments but also use in this study protein-ligand interactions for a variety of applications. Undergraduate students that are in the NSF funded S-STEM program have been trained in these techniques. This technique allows us to map the ligand protein interactions.
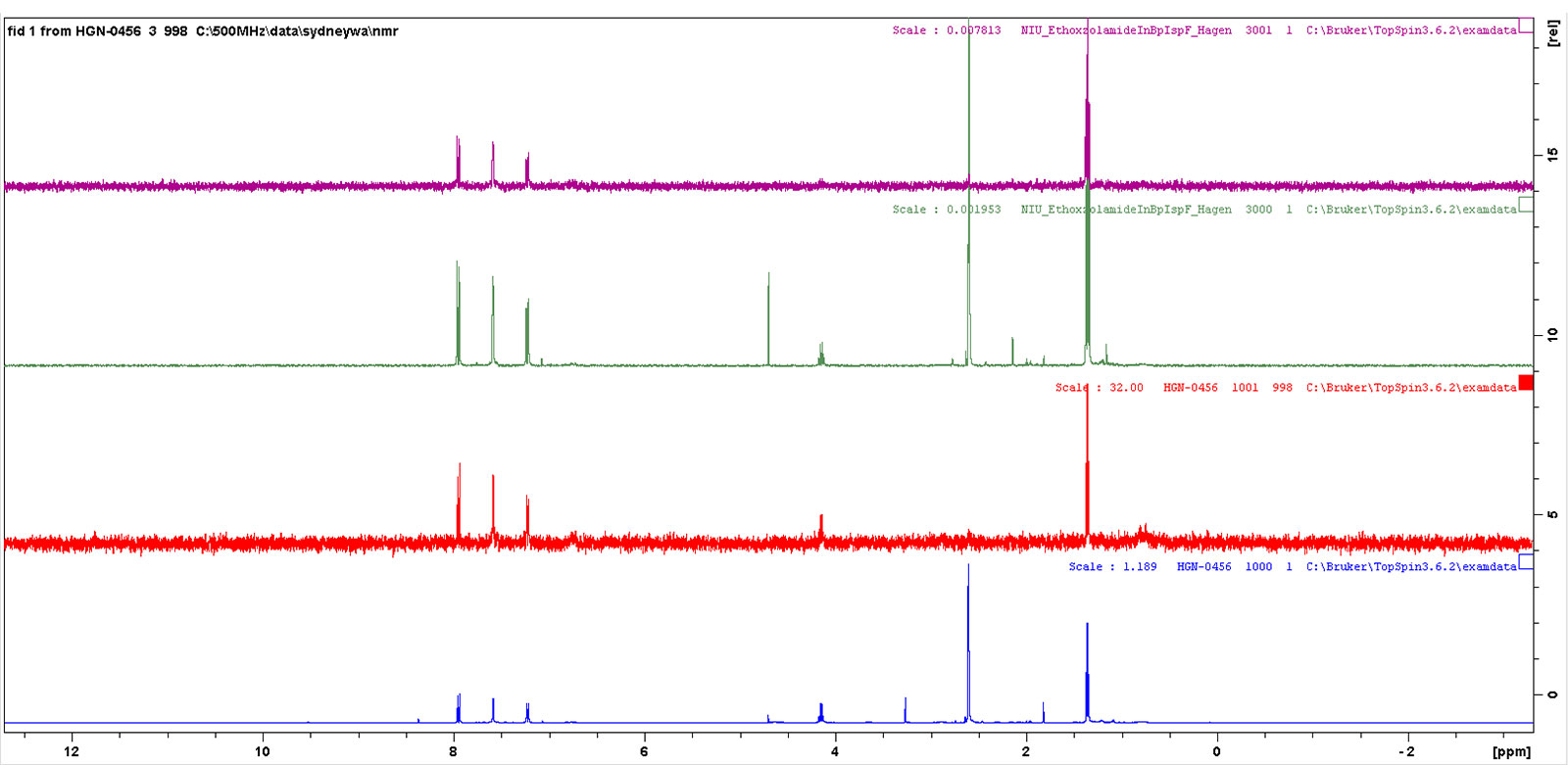
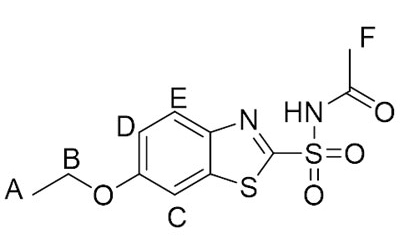
| Proton | Relative Percent Saturation Transfer |
|---|---|
| A | 14-19% |
| B | 32-39% |
| C | 100% |
| D | 31-37% |
| E | 42-44% |
| F | Negative-5% |
Design and Synthesis of Novel Small‐molecules To Selectively Inhibit the Phosphodiesterase Type 4 (PDE4) Enzymes
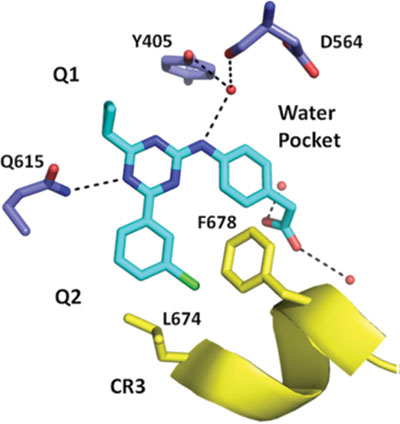
Professor Hagen has a longstanding interest in phosphodiesterase (PDE) inhibitors and has ground-breaking publications and patents in this area. Phosphodiesterases hydrolyze cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP) to their cognate 5’-monophosphate derivatives. There are eleven different PDE superfamily members (PDE1-11) and inhibitors have been developed to prolong the effects of physiological processes mediated by cAMP or cGMP. Phosphodiesterase 4 (PDE4) is the primary enzyme that regulates the turnover of cAMP. PDE4 is comprised of four genes (PDE4A-D). We have synthesized selective inhibitors for the PDE4D and PDE4B enzymes and have collaborations that have resulted in numerous crystal structures that have provided insight into the basis for PDE4D and PDE4B selectivity.
Natural Product Isolation and Characterization
The Hagen group has a collaboration with Kenyatta University to isolate and characterize natural products from plants used in traditional medicine. For centuries, plants have been used for the treatment of various diseases and they can be investigated for the discovery of new therapeutic agents. In many developing countries such as Kenya, the use of medicinal plants for the treatment of various ailments is quite common. It is estimated that about 70% of the Kenyan population uses medicinal plants for primary health care. Divergent Kenyan communities like Ogiek, Taita, and Maasai use medicinal plants as therapeutics since they live far away from modern medical facilities. In the Ogiek community, 96 % of the population uses medicinal plants as their major therapeutic agents. Various studies have reported the antimicrobial properties and efficacy of Kenyan medicinal plants. Evaluation of the medicinal properties of such plants and isolation of the phytochemicals can lead to the discovery and isolation of new therapeutic agents for the treatment of a variety is diseases.
| Family Name | Botanical Name | Common Name | Part Used | Preparation | Traditional Use |
|---|---|---|---|---|---|
| Asteraceae | Artemisia annua | Sweet wormwood | Leaf, flower | Decoction | Malaria, Psoriasis, Infections |
| Lamiaceae | Ajuga remota | Bugleweed | Leaf | Decoction | Malaria, Chest pains |
| Solanaceae | Physalis peruviana | Cape gooseberry | Leaf | Decoction | Typhoid, Pneumonia |
| Rosaceae | Prunus africana | Red stinkwood |
Stem bark |
Decoction | Pneumonia/chest pain, loss of appetite |
| Boraginaceae | Cordia africana | Giku | Bark, leaf | Decoction | Fatigue, anti-inflammatory |
| Fabaceae | Senna didimobotrya | Candelabra tree | Leaf | Decoction, steam | Pneumonia |
| Euphorbiacece | Bridelia micrantha | Mitzeeeri | Stem bark | Chew | Chest pains |






